|
|
 |
WinXP program by Thomas A. Germer to compute integration of the bidirectional reflectance distribution function (BRDF) over a given solid angles for defects on a plane surface.
FORTRAN program listed in B.R. Johnson: Light Scattering from a Spherical Particle on a Conducting Plane: Part I. Normal Incidence, Aerospace Tech. Rep. TR-0091(6945- 08), 1991, Space and Environment Technology Center, The Aerospace Corporation, P.O. Box 92957, Los Angeles CA 90009
C++ object class library (SCATMECH) to disseminate simple models for light scattering from smooth surfaces. The library includes routines for scattering from spherical particles (Rayleigh, Rayleigh-Gans, and Mie) and routines for various models of scattering from surfaces. Data types for manipulating different representations for the polarization of light and the polarimetric properties of materials,specifically Mueller matrices, Stokes vectors, and Jones matrices and vectors.
Open-source Lattice Boltzmann Method solver for electromagnetic wave scattering and radiation force calculations.

Program written by Gorden Videen and modified by Dat Ngo. The program follows the derivation of Videen and Ngo given Videen and Ngo, J Opt. Soc. Am. A 14, p. 70 (1997).
Fortran code to compute the field of an fith oder Gaussian beam
Program written by Gorden Videen and modified by Dat Ngo. The program follows the derivation of Gorden Videen given in his dissertation and the articles Videen, Opt. Comm. 115, p. 1 (1995); Videen, JOSA A 8, p. 483 (1991); 9, p. 844 (1992).
- Link (local copy, 25 Feb 1998)
Function package for calculating light transport in multiple scattering media.
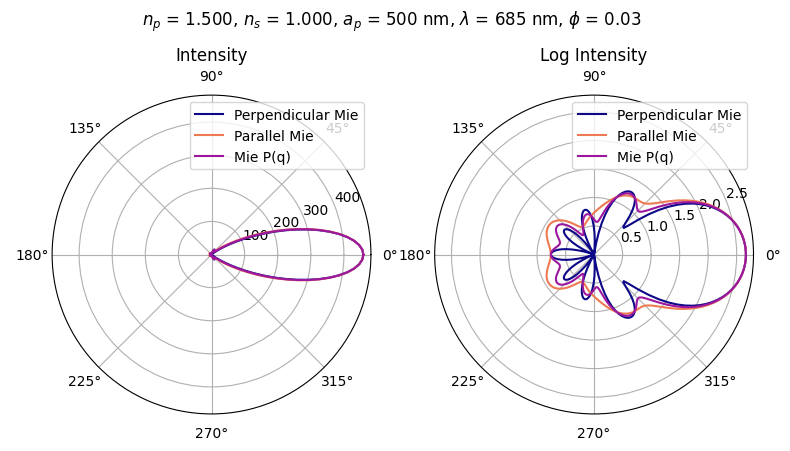
FORTRAN program listed in Gorden Wayne Videen: Light scattering from a sphere on or near an interface, Ph. D. 1992, University of Arizona; UMI Order Number 9220701
A vector finite element method for electromagnetic scattering (C++).

|
|
 |
|